- Reproduction
- bacteria reproduce asexually by fission
- circular DNA replicates and is separated via a membrane attached complex
- cell elongates and divides into two cells
- process does not take long (20' minimum) and produces a clone of organisms
- Prokaryotes have inheritable traits
- bacteriophage attach to receptors in cell wall
- mutations alter the structure of the receptors and bacteriophage cannot attach so bacteria become"resistant" to infection
- bacteria can mutate to
- become resistant to specific antibiotics
- be unable to synthesize specific amino acids
- be unable to utilize specific sugars for energy
- etc.
- Sexual reproduction in bacteria
- Joshua Lederberg and Ed Tatum demonstrated that after mixing two, distinguishable strains of bacteria together, they could isolate recombinants between them at a very low frequency
- one strain was met-bio-thr+leu+
- the other strain was met+bio+thr-leu-
- and the recovered recombinants were not mutant for all four traits or met+bio+thr+leu+
- others demonstrated that bacteria can be categorized as either male or female depending on whether they have an extra piece of DNA or not
- the extra piece of DNA (known as a plasmid) replicates separately from the bacterial chromosome
- it contains genes that promote the transfer of the DNA from one cell to another including
- information to produce a tube or pilus that connects two cells together and through which the plasmid DNA is transmitted
- a bacterium containing the fertility plasmid or F plasmid is classified as a male (F+) and can transmit the plasmid to other bacteria (F-)
- Jacob and Wollman investigated the transfer of chromosomal genes in bacteria
- depends on a few of the F+ bacteria in which the F plasmid has become integrated into the bacterial chromosome
- the bacterial chromosome is transmitted as if part of the plasmid but the entire chromosome is transmitted first
- for a particular strain, the bacterial chromosome is always transmitted from the same point and in the same direction
- the transfer of genetic material from one bacterium to another is very sensitive to shear so that matings can be easily interrupted
- the transfer of DNA from one cell to the other is actually driven by DNA synthesis
- recombination within the recipient involves incorporation of donor DNA into recipient DNA via a crossing over process
- Earlier experiments of Griffith, Hotchkiss, and Avery and co-workers showed that bacteria could take up DNA from the environment and become transformed; this process can occur in nature
- Some bacteriophage are able to transfer bacterial DNA from a previous host cell to a newly infected cell in a process called transduction.
- Some bacteria contain other DNA elements similar to the F factor; these are called episomes
- One class of episomes are the R factors or Resistance Transfer Factors
- carry gene or genes conferring resistance to one or more antibiotics
- can be transferred from one strain to another upon contact
- can pick up genes to become resistant to multiple antibiotics all at once
- a serious hospital problem
- Transposable elements move DNA by the action of enzymes that cut and splice DNA
- Regulation of Gene Expression I
- it is important that bacteria regulate the expression of their genes in the interest of efficiency
- must be able to turn genes on or off when the gene products are needed or not needed
- must have a system for sensing the presence or absence of metabolites
- and must be able to convert that information into control of mRNA transcription
- some genes are never regulated other than by the efficiency of promoter binding
the products of these genes are said to be constitutive
- other gene products appear only when their substrate molecules are present (degradative pathways)
the products of these genes are said to be inducible
- and other gene products stop being synthesized when their end products are present at a significant concentration (synthetic pathways)
the products of these genes are said to be repressible
- Regulation of Gene Expression II: the lac operon
- Jacob, Monod, and Wollman mapped the genes associated with the utilization of lactose and found that they mapped very close to each other
- 3 genes code for enzymes:
-
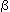 -galactosidase hydrolyzes the bond between the galactose and the glucose of lactose
-galactosidase hydrolyzes the bond between the galactose and the glucose of lactose
- permease makes the uptake of
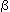 -galactosides easier
-galactosides easier
- transacetylase
- Jacob, Monod and Lwoff found that the three genes are co-ordinately regulated
- if lactose is not present, none of the gene products are present
- if lactose is present as the sole sugar source, the gene products appear in near equi-molar amounts
- if lactose is removed as the sole sugar source, new gene products are no longer made and previously made enzymes decay
- Jacob, Monod and Lwoff found mutants that confounded the simple regulation rules
- mutants located in the region between the promoter and the first gene turned all three genes in to constitutive synthesis:
these mutants defined the operator region of the gene which gives the entire gene set its name as an operon
- another set of mutants located near the operon but not in it made the operon non-inducible:
these mutants defined the repressor region
- additional mutants of the repressor also made the operon constitutive
- an animation of the lac operon
- basic model of lactose operon
- the lac repressor gene is read by RNA pol as a constitutive gene and lac repressor is made at all times at a low level
- the repressor protein (as a tetramer) binds to the DNA sequences in the operator segment of the lac operon
- when repressor is bound to the operator, RNA pol cannot bind to the lac promoter and transcribe the lac operon
- although the affinity between repressor and operator is high, repressor does release from the operator at times which allows a very low level of"escape" synthesis
- the disaccharide lactose can operate as an inducer and acts by binding to repressor
- the repressor protein is allosteric and the bound lactose causes a conformational change with does not allow binding with the operator
- thus, in the presence of lactose, the repressor molecules are titrated into the inactive state
- once the operator is not covered by repressor, RNA pol can bind to the lac promoter and transcribe the operon
- this results in the translation of the mRNA and the production of
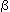 -galactosidase which cleaves lactose into glucose and galactose for energy consumption
-galactosidase which cleaves lactose into glucose and galactose for energy consumption
- as the lactose is used up and its concentration decreases, lactose is released from repressor
- repressor without bound lactose is able to bind to the operator and stop RNA pol from binding at the promoter
- taking us back to the beginning
- the mutants that defined the system
- O C or operator constitutive mutants are changes in the DNA sequence of the operator that is not recognized by the repressor so it doesn't bind and RNA pol can read the operon at will
- I S or super-repressor mutants had no affinity for lactose and could not end up in a conformation that would not bind the operator
- I - mutants either do not make repressor or make a repressor that does not recognize the DNA sequences of the operator which results in constitutive expression of the operon
- Jacob, Monod, and Lwoff received the Nobel Prize for this work
- Regulation of Gene Expression III: the tryp operon
- the tryp operon consists of 5 co-ordinately expressed structural genes controlled by a promoter and operator with regulation by a constitutively expressed repressor gene nearby
- however, the end product of the tryptophan synthetic pathway (tryptophan) acts as a co-repressor in the regulation of gene expression
- when tryptophan is abundant, tryptophan binds to repressor which allows the complex to bind to the operator; tryptophan acts as a co-repressor
- when tryptophan concentration becomes low, the repressor loses its bound tryptophan and can no longer bind to the operator
- then the RNA pol can bind at the promoter and transcribe the tryp operon
- thus, the tryp operon is repressible
- Regulation of Gene Expression IV: Positive Regulation
- the lac and tryp operons show negative regulation through operator binding to prevent RNA pol binding at the promoter
- the lac operon also shows positive regulation since it is one of several operons showing catablite repression
- glucose is the preferred sugar for energy metabolism but when glucose is absent, the cell will utilize any available source of energy
- the signal that glucose is in low supply is a rise in the concentration of cAMP
- a cAMP receptor protein binds cAMP to form the CRP-cAMP complex
- the CRP-cAMP complex binds to the promoter/operator region of all operons involved in sugar metabolism
- this binding enhances the binding of RNA pol at the promoter and the reading of the operon
- thus, the cell activates the various enzyme systems that can utilize most any energy source that the cell encounters
- this is a positive regulation system to enhance transcription
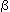 -galactosidase hydrolyzes the bond between the galactose and the glucose of lactose
-galactosidase hydrolyzes the bond between the galactose and the glucose of lactose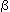 -galactosides easier
-galactosides easier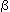 -galactosidase which cleaves lactose into glucose and galactose for energy consumption
-galactosidase which cleaves lactose into glucose and galactose for energy consumption